YING YANG
I am a Ph.D. candidate in Scientific Computing, and Molecular, Cellular and Developmental Biology (a joint doctoral program hold by Molecular, Cellular, and Development Biology Department and Michigan Institute for Computational Discovery and Engineering) at the University of Michigan, supervised by Prof. Daniel J. Klionsky. My current research interest focuses on deploying deep learning models and numerical modeling to address real-world challenges, with a particular emphasis on biomedical contexts. I am enthusiastic about tackling these challenges and leveraging AI to revolutionize scientific research.
As an addition to my doctoral studies, I obtained my Master of Science degree in in Computer Science and Engineering from Electrical Engineering and Computer Science department at the Univerisity of Michigan, and my Master of Science degree in Bioinformatics from Medical School at the Univerisity of Michigan.
Before Michigan, I received my bachelor's degree from Fudan University, Shanghai.
News: I started my internship at Google on 4/1/2024 (Not an April Fools Hoax)!
Email Google Scholar Github LinkedIn Instagram

Education

Master of Science in Computer Science and Engineering
GPA: 4.0/4.0
- EECS 592 Artificial Intelligence Foundations A
- EECS 545 Machine Learning A
- EECS 586 Design and Analysis of Algorithms A+
- EECS 587 Parallel Computing A
- EECS 568 Mobile Robotics A
- EECS 583 Advanced Compilers
- EECS 598 Special Topics: Deep Learning
- EECS 598 Special Topics: Artificial General Intelligence

Master of Science in Bioinformatics
GPA: 4.0/4.0
- BIOINF 527 Introduction to Bioinformatics and Computational Biology A+
- MATH 425 Probability A
- BIOINF 575 Programming Laboratory in Bioinformatics A+
- BIOINF 545 High-throughput Molecular Genomic and Epigenomic Data Analysis A+
- BIOSTAT 521 Applied Biostatistics A
- BIOINF 580 Introduction to Signal Processing and Machine Learning in Biomedical Sciences

Ph.D. in Molecular, Cellular and Developmental Biology, and Scientific Computing
GPA: 4.0/4.0
Advisor: Prof. Daniel J. Klionsky
Research Interest: Autophagy, Cancer Genomics, Machine Learning, etc.

Bachelor of Science in Biological Sciences
National Top Talent Undergraduate Fellow
Research Thesis: The novel role of the Golgi protein Otg1 in hormone secretion and glucose homeostasis. (Supported by the Undergraduate Research Opportunity Program)
Advisor: Prof. Xiaohui Wu, Prof. Min Han and Prof. Tian Xu
Publications
Total citations: 12262; h-index: 10.
Citations as first, second and third author: 572.
Updated on 03/01/2023.
A novel role of ATG9A and RB1CC1/FIP200 in mediating cell-death checkpoints to repress TNF cytotoxicity
Ying Yang, Daniel J. Klionsky
Autophagy, 2023
Scheduling Hyperparameters to Improve Generalization: From Centralized SGD to Asynchronous SGD
Jianhui Sun, Ying Yang, Guangxu Xun, Aidong Zhang
ACM Transactions on Knowledge Discovery from Data (TKDD), 2023
Follicular lymphoma-associated mutations in the V-ATPase chaperone VMA21 activate autophagy creating a targetable dependency
Fangyang Wang, Ying Yang, Gabriel Boudagh, Eeva-Liisa Eskelinen, Daniel J. Klionsky, Sami N. Malek
Autophagy, 2022
Mutations in V-ATPase in follicular lymphoma activate autophagic flux creating a targetable dependency
Fangyang Wang, Ying Yang, Daniel J. Klionsky, Sami N. Malek
Autophagy, 2022
Follicular lymphoma-associated mutations in the V-ATPase chaperone Vma21 activate autophagy by dysfunctional V-ATPase assembly
Ying Yang, Zhihai Zhang, Daniel J. Klionsky
Autophagy Reports, 2022
Upstream open reading frames mediate autophagy-related protein translation
Ying Yang, Damián Gatica, Xu Liu, Runliu Wu, Rui Kang, Daolin Tang, Daniel J. Klionsky
Autophagy, 2022
Downregulation of autophagy by Met30-mediated Atg9 ubiquitination
Yuchen Feng, Aileen R Ariosa, Ying Yang, Zehan Hu, Jörn Dengjel, Daniel J. Klionsky
Proceedings of the National Academy of Sciences (PNAS), 2021
A perspective on the role of autophagy in cancer
Aileen R Ariosa*, Vikramjit Lahiri*, Yuchen Lei*, Ying Yang*, Zhangyuan Yin*, Zhihai Zhang*, Daniel J. Klionsky (*equal contribution)
Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 2021
An AMPK-ULK1-PIKFYVE signaling axis for PtdIns5P-dependent autophagy regulation upon glucose starvation
Ying Yang, Daniel J. Klionsky
Autophagy, 2021
A Stagewise Hyperparameter Scheduler to Improve Generalization
Jianhui Sun, Ying Yang, Guangxu Xun, Aidong Zhang
Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining (KDD), 2021
Moments in autophagy and disease: Past and present
Xin Wen*, Ying Yang*, Daniel J. Klionsky (*equal contribution)
Molecular Aspects of Medicine, 2021
Follicular Lymphoma-associated mutations in the V-ATPase assembly factor VMA21 activate autophagic flux
Fangyang Wang, Ying Yang, Gabriel Boudagh, Eeva-Liisa Eskelinen, Daniel J. Klionsky, Sami N. Malek
Cancer Research, 2021
A novel reticulophagy receptor, Epr1: a bridge between the phagophore protein Atg8 and ER transmembrane VAP proteins
Ying Yang, Daniel J. Klionsky
Autophagy, 2021
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)
Daniel J. Klionsky, ..., Ying Yang, et al.
Autophagy, 2021
LRRC8 family proteins within lysosomes regulate cellular osmoregulation and enhance cell survival to multiple physiological stresses
Ping Li, Meiqin Hu, Ce Wang, Xinghua Feng, ZhuangZhuang Zhao, Ying Yang, Nirakar Sahoo, Mingxue Gu, Yexin Yang, Shiyu Xiao, Rajan Sah, Timothy L Cover, Janet Chou, Raif Geha, Fernando Benavides, Richard I Hume, Haoxing Xu
Autophagy, 2021
Proceedings of the National Academy of Sciences (PNAS), 2020
The roles of ubiquitin in mediating autophagy
Zhangyuan Yin*, Hana Popelka*, Yuchen Lei*, Ying Yang*, Daniel J. Klionsky (*equal contribution)
Cells, 2020
Autophagy and disease: unanswered questions
Ying Yang, Daniel J. Klionsky
Cell Death & Differentiation, 2020
A novel role of UBQLNs (ubiquilins) in regulating autophagy, MTOR signaling and v-ATPase function
Ying Yang*, Daniel J. Klionsky
Autophagy, 2020
PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex
Gonen Memisoglu, Vinay V. Eapen, Ying Yang, Daniel J. Klionsky, James E. Haber
Proceedings of the National Academy of Sciences (PNAS), 2019
Disruption of the Golgi protein Otg1 gene causes defective hormone secretion and aberrant glucose homeostasis in mice
Guangxue Wang*, Rongbo Li*, Ying Yang*, Liang Cai, Sheng Ding, Tian Xu, Min Han, Xiaohui Wu (*equal contribution)
Cell & Bioscience, 2016
Selected Research Themes

- We introduced a genomics discovery framework that uses graph embeddings derived from knowledge graphs as generalized representations of genomics data.
- The discovery framework consists of a joint scheme that first recalls relevant candidates, and next ranks the candidates based on multiple target pathways/diseases.
- The image shows the schematic workflow to screen candidate genes including constructing KG, representation learning, recalling candidates, and ML prediction for ranking.
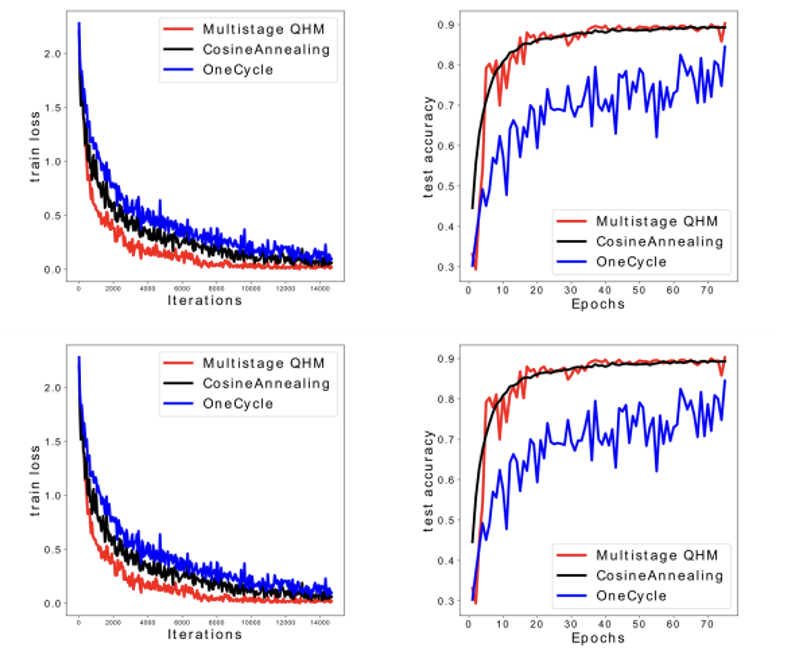
- We showed theoretically how hyperparameter configurations in Stochastic Gradient Descent Momentum connect to the generalization of optimizers.
- We optimized the hyperparameters in a principled fashion.
- We improved over existing auto-tuners on image classification tasks.
- The image shows the testing curves and accuracy with various learning rate schedulers and optimizers.
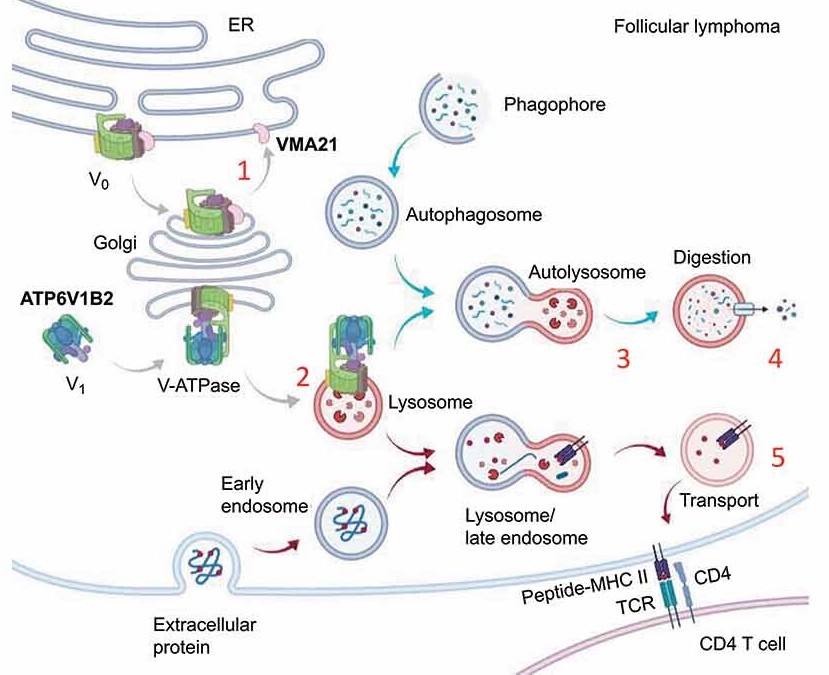
- We identified mutations in ATP6AP1, ATP6V1B2 and VMA21 in a combined 30% of follicular lymphoma (FL), together constituting a major novel mutated pathway in this disease.
- We characterized the molecular pathologies underlying the FL mutations.
- We proposed a novel therapeutic opportunity for FL patients by targeting autophagy alone or in combination with other targeted therapies.
- The image shows the Pathobiology of VMA21 p.93X mutations.
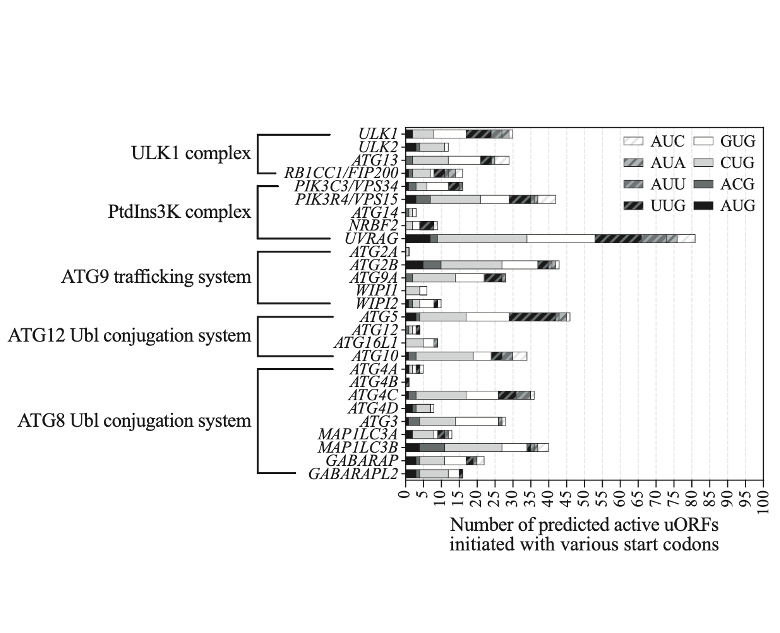
- We identified the upstream open reading frame (uORF)-mediated translational control of multiple Atg proteins in Saccharomyces cerevisiae and in human cells.
- We found translational events of autophagy-related genes mediated by the uORF is dependent on the surrounding contexts of the uORF start codon, and on the Gcn2 kinase.
- We delineated a series of genes encoding the core autophagy machinery with active uORFs in human cells.
- The image shows the set of core autophagy machinery with uORFs as translational control elements.
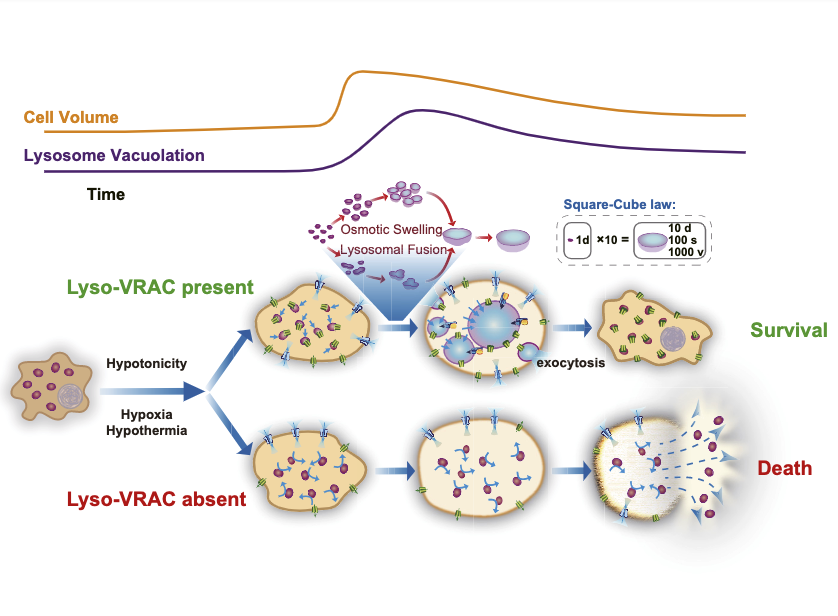
- We showed that intracellular LRRC8 proteins acting within lysosomes play an essential role in cellular osmoregulation.
- We found Lyso-VRACs allow lysosomes of mammalian cells to act as the cell's bladder.
- We identified that Lyso-VRACs is essential in enabling cells to mount successful homeostatic responses to multiple stressors.
- The image shows the model for how Lyso-VRAC–mediated lysosome osmoregulation behavior is protective for cell survival.
Services
Peer Reviewer for Journals & Conferences
MLGenX @ ICLR 2024 (International Conference on Learning Representations)
International Conference on Machine Learning
Aging
Annals of Medicine
Autophagy
Autophagy Reports
Cell & Bioscience
Frontiers in Genetics
International Journal of Computational Biology & Drug Design
Molecular & Cellular Biochemistry
Pathology & Oncology Research
Instructor / Mentor
Graduate Student Instructor
Instructed 270+ undergraduate students
Graduate Research Mentor
Instructed two rotation graduate students in the Klionsky lab
Awards
Conference on Neural Information Processing Systems Travel Award 2023
Knowledge Discovery and Data Mining Travel Award
Rackham Research Grant
Okkelberg Fellowship
Fun Facts
- The first "surprise" image on my website is my favorite photo for my first cat - Ginger Ale. He gets this name because of his fur color.
- The second "surprise" image on my website is my second furry friend, Fluffy. It is obvious why he gets this name. Every time we visit the vet, he always gets the praise that "HE IS FLUFFY".